Tools for modern genomics
Developing approaches to make sense of cutting-edge DNA/RNA sequencing data

Technology for DNA sequencing has dramatically changed over the past two decades. Now, bacterial genomes can be sequenced for less than the cost of a smartphone, and DNA and RNA sequencing technology is being applied in an increasing number of settings around the world. From giving us a new window to study the ecology of Earth’s ecosystems to having new tools for industrial applications in the food and energy sectors, DNA sequencing technology is revolutionizing our ability to understand and interact with biological systems.
With the rapid progression of DNA sequencing technology comes rapid changes in data analysis needs and data analysis volumes. Bioinformatics tools written a few years ago can sometimes find themselves obsolete due to the development of new computational approaches, and increasingly, software engineering approaches are needed to manage the high amounts of sequencing data that are stored and analyzed by research groups. Higher volumes of data also create new opportunities to perform meta-analyses of existing DNA data through comparative genomics or environmental surveys. One of the ongoing themes of my work is to develop new approaches and workflows to analyze modern DNA/RNA sequencing data, with the hope that these workflows can contribute to making cutting-edge DNA/RNA sequencing data more accessible to the research community in the long-term.
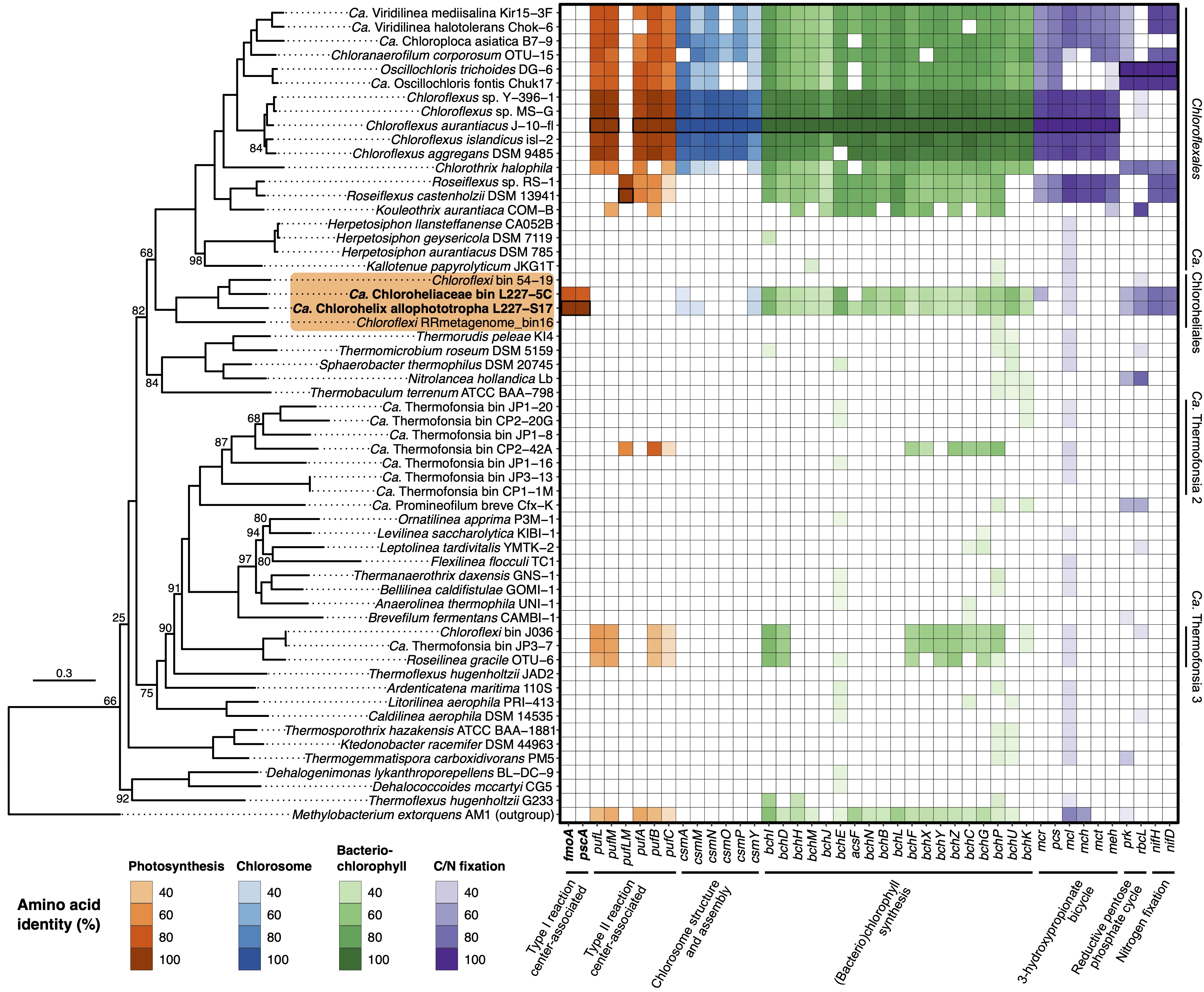
My work in this research theme is typically driven by my own data analysis needs. To address an interesting research question, often bioinformatics approaches are needed that don’t quite exist yet or are not yet engineered to tackle high volumes of input data. Over the past several years, I have helped develop semi-automated visualization tools for ‘omics data that help make sense of high volumes of underlying sequence information (see figure below for an example). My current work is to develop a hybrid long- and short-read sequence assembly pipeline for genomes with circular DNA elements, called Rotary, that uses best practices to avoid common problems that occur around the start point of circular genomes in FastA files. Beyond this, I am also working on several “big data” projects to either analyze environmental DNA/RNA data from field surveys or to do global biogeography based on publicly available and archived sequence data. Look forward to sharing the results of these works soon - a couple are already available online, such as Tsuji et al., ISME J, 2020 and Tsuji et al., Nature, 2024.